X
X
Leaving Community
Are you sure you want to leave this community? Leaving the community will revoke any permissions you have been granted in this community.
No
Yes
X
Do you know what you don't know? A gap analysis of Neuroscience Data.
Do you know what you don't know? A gap analysis of Neuroscience Data.
My thesis adviser, a colorful spirit and one whose wisdom will long be missed, used to say that undergraduate or professional students differed from graduate students in that they were asked to learn what was known about a subject, while graduate students were asked to tackle the unknown.We, in higher education, are essentially seeking to find out what is not known and start to come up with new answers. How does one find out what is not known? In fact, is it possible to do that? Don't most graduate students or postdocs add onto a lab's existing body of knowledge? Adding to the unknown by building on the known? If this is how we work then does this create a very skewed version of the brain? How would we even know what is truly unknown?
Now we enter the omics era, where we try to find out all things about a set of things. We no longer want to know about a gene, we want to know about all of the genes, the genome of an organism. We want to account for all things of the type DNA and figure out which parts do what. In neuroscience, this tends to be a little more difficult. Mainly because we do not have a finite list of things that we can account for. We have a large quantity of species with brains, or at least ganglia, we have billions of cells and many more connections between them in a single human brain. The worst part is that these connections are not even static so a wiring diagram is only good for a few minutes for a single brain and then the brain reorganizes some of these connections.
Is the hope for an "omics" approach to neuroscience?
Well, the space is not infinite and has been studied over the last 100+ years so we have some ways of getting at the problem. We have a map!
Can we use this map to figure out some basic information about what we do and do not study? Well, the short answer at least for some things seems to be yes!
The Neuroscience Information Framework (neuinfo.org) project has been aggregating data of various sorts that is useful to neuroscientists, and also a set of vocabularies for all of the brain parts, the map of the nervous system. So we can start to look at which labels are used for tagging data, and which are found in the literature? Are all parts of the brain equally represented by relatively even amounts of data or papers or are there hot spots and cold spots for data?
Below is a heat map generated using the Kepler tool for data sources vs brain regions across the canonical brain regions (a hierarchy built to resemble what one may find in a graduate level text book of neuroanatomy).
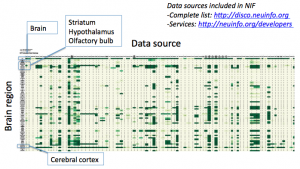
Albeit the heat map is very hard to read (the darker the green the more data, you can generate your own by clicking on the graph icon in NIF), there is little doubt that all brain regions are not equal, and some have very little data, while others have a plethora of data begging the question: Are there popular brain regions and not-so-popular brain regions?
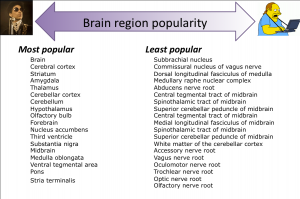
Indeed, there are brain region annotations that are found more often, when looking at data and much like pop stars, they tend to have shorter names. The most popular data label is actually brain, and the least popular appears to be the Oculomotor nerve root. This is starting to tell us that most data is just labeled as "brain vs kidney", but can we do better as neuroscientists? In fact, we can break down the labels into major regions like hindbrain, midbrain and forebrain and add up all of the data that fit into each of these. Note most of the data are attributed to the forebrain, housing some of the most popular brain regions such as the cerebral cortex and the hippocampus, but the hindbrain also comes back with some reasonable data, mainly for the cerebellum. It turns out that adding up all the data labels for midbrain regions results in an awkward sense that the midbrain may be completely non-essential to brain research. On the other hand, removing the midbrain appears to be essential to life, so why do neuroscientists not know much or at least publish much about the midbrain?
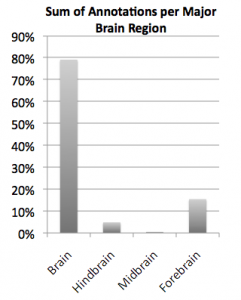
So if you are hiding a big pile of data about the midbrain in your desk drawer, I would like to formally ask you to share it with NIF (just email info@neuinfo.org) so that I can stop thinking of the midbrain as the tissue equivalent of fly-over country.
X









