X
X
Leaving Community
Are you sure you want to leave this community? Leaving the community will revoke any permissions you have been granted in this community.
No
Yes
X
The Factorial Problem in the Annotation of the Scientific Paper
The Factorial Problem in the Annotation of the Scientific Paper
For the better part of the last 200 years, scientists and researchers have been honing the process of publishing scientific articles by carefully citing the people and ideas leading up to their own thinking, they have refined how they speak about their own discoveries, and they have become much more prolific as a community. Indeed, the “publish or perish” mantra has led to an unprecedented proliferation of publishable material, so much so that no scientist can reasonably expect to read everything in his or her field.We have just heard at the Beyond the PDF conference and echoed throughout the scientific community that the scientific paper should have live links to data, videos and software tools so researchers can almost, practically, recreate experiments. The scientific paper should have a provenance of scientific discourse, i.e., who said what about whom. The scientific paper should really not be called a paper at all; it should be called a publication and should never again be thought of as a flat object.
Okay, great, all these new dimensions will then become exactly that, orthogonal axes to the text, and there will be references to all these wonderful and useful things. However, authors and authoring tool builders may begin a bit of a backlash, if they have not done so already. Why? Well, let’s just consider the standard paper as using n repositories of data, consisting of 5 vendors for materials, 1 database where data are stored that the paper was based on, 3 software tool pointers at different open and closed source repositories with which the data were analyzed, running on some well-defined platform, and a list of 4 ontologies which were used to annotate the various important parts of the paper. Now, consider that the author or toolmaker would need to access these repositories of data, software tools, vendors and ontologies to pull the unique identifiers that allow text-mining systems to “read” the paper. That is fine; in this example, the total number of databases that would be accessed is only 13-16, depending on whether we want to consider system emulation as part of our parameters.
In astronomy, which runs entirely on a few very large datasets accessible by everyone in the community, this problem is solved. The community sets standards for scientific discourse, Pluto be damned! Everyone links to everyone they care to link to, and all communication issues can be boiled down to whether the astronomers are talking to other astronomers or to journalists about to spin an astrology piece pointing to some new zodiac sign.
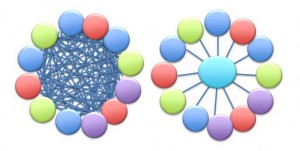
So back to our non-astronomy science paper above, let us now consider that each of the parties involved would need to link to each of the other parties involved. For example, the mouse provider would like to say that this article discusses some new discovery based on their knockout mouse, the software tools would like to point to datasets useful for testing data, ontologies would like to capture new instances, etc. If we take this to the extreme, we are faced with a factorial (n!) connection between the various players in the scientific publication process. Now let’s consider that data can be stored in one of 2061 databases useful to neuroscience, and we see that a rather large number of the connections emerge. The problem of connectivity becomes a large tangled “hairball” from which no answer can come other than “this is too darn complicated.”
Caption: hairball (n!) vs. one link solution (n+1)
So how do we disentangle the “hairball” and let the tools that people produce help scientists write new research publications (not flat papers and not tangled hairballs)? Here is one possible answer: Create a platform which the whole community can point to, which can route the appropriate links to the appropriate place, reducing the overhead of “everyone linking to everyone” (n!) down to a manageable “everyone linking to one” (n+1) problem?
That seems like a useful exercise, and an exercise that is emerging as a theme in the shared names, DataCite and NIF projects.
X