X
X
Leaving Community
Are you sure you want to leave this community? Leaving the community will revoke any permissions you have been granted in this community.
No
Yes
X
RRID's are in the wild! Thanks to JCN and PeerJ
RRID's are in the wild! Thanks to JCN and PeerJ
We believe that reproducing science starts with being able to know what "materials" were used in generating the results.Along with a truly dedicated group of volunteers from academia, government and non-government institutes, publishers and commercial antibody companies we have been running the Resource Identification Initiative (RII).
This initiative is meant to accomplish the following lofty goal: Ask authors to uniquely identify their antibodies (no easy task), organisms (an even harder task), and the databases and software tools that they used in their paper.
In order to ask them at the appropriate time, we gathered a group of journal chief editors to help us ask this question when authors are most interested in answering the question during the process of publication. We created many things to help them identify these things such as a database that stores information for 5 of the most common species used in experiments, antibody catalogs from over 200 vendors, and a database and tool catalog that contains over 3000 software tools and over 2500 academic databases, the largest of its' kind.
We have been granted 3 months to determine if authors would actually do this. It has been two months, we fielded requests from about 30 users who could not find their resources, there have been more than 40 new software tools or databases registered to our tools registry, and more than 100 antibodies, but we kept waiting for RRIDs to show up in the literature.
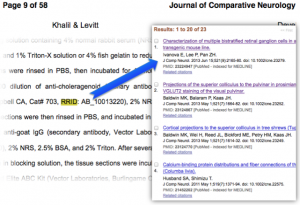
Today our wait is over thanks to two papers, Khalil and Levitt in the Journal of Comparative Neurology and Joshi et al in PeerJ.
These authors apparently were able to correctly identify resources such as Matlab, NeuroLucida, ProteinDataBank and antibodies including anti-cholera toxin antibody from List Bio.
What does this tell us?
Well to start that this process is not impossible! That identifiers do exist for many things or the process of obtaining new ones is not so difficult that people can't do this. It also tells us that when asked at the right time, authors are willing to go the extra step, find and provide identifiers to their reagents or software tools!
Great, but why do I care about a single paper that uses an antibody or Matlab?
Well, it turns out that for many years JCN and NIF staff have been working diligently to link papers through that same identifier so in the case of this cholera toxin antibody we have marked 23 other papers that have used it since 2006.
X









